
cpout/Object relationships.csv: contains neighbor information in form of an edge list between cells. channel intensities) and acquisition metadata. cpout/Image.csv: contains image-level measurements (e.g.
CELLPROFILER CSV DOWNLOAD
The results will be stored in a folder named “output_directory”.Ĭlick here to download the script to load images from PMA. cpout/Experiment.csv: contains metadata related to the CellProfiler version used. This will start CellProfiler in headless mode and execute “my_pipe_line.cppipe” against the image we fetched. Next we invoke CellProfiler, provided that we have our pipeline ready, in the following manner: CellProfiler.exe -c -r -data-file input.csv -o /output_directory -p my_pipe_line.cppipe It will also create a CSV file named input.csv, again in the current directory.

Keep track of dependencies of CellProfiler Analyses - first run an illumination correction and then your analysis. As a non exhaustive list you need to: Trigger CellProfiler Analyses, either from a LIMS system, by watching a filesystem, or some other process. For this example, we will be using the demonstration pipeline accessible from the Welcome screen in. If you are running a High Content Screening Pipeline you probably have a lot of moving pieces. Run your CellProfiler pipeline or project and save the per-image or per-image measurement output as a comma-delimited text file (CSV) using the.
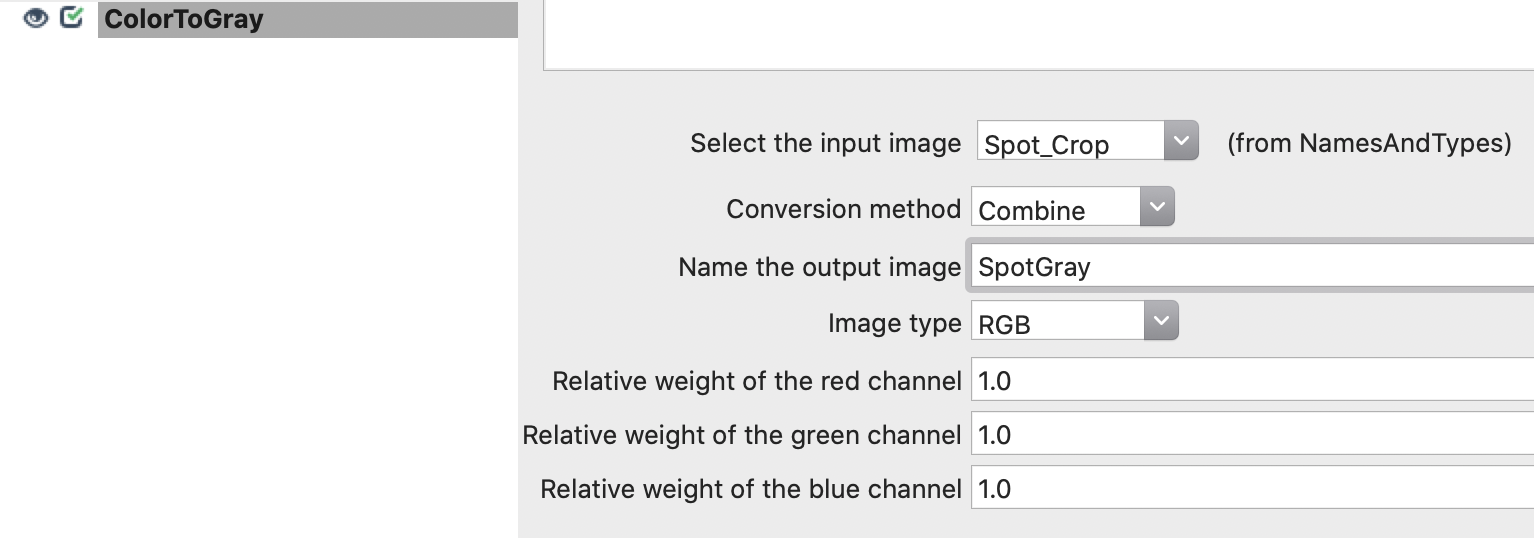
CELLPROFILER CSV HOW TO
This will fetch a snapshot of the image and save it as slide1.jpg in the current directory. How to use CellProfiler 2.1 output in Spotfire DecisionSite. A simple case for an RGB image is to print the path to the image file inside the CSV.įor example if we want to feed the image at c:slidesslide1.svs to CellProfiler, we first fetch a snapshot using the attached Python program by issuing: python CellProfiler.py c: /slides/slide1.svs To achieve this, first we have to download an image from PMA.start and create an accompanying CSV file for it that contains required meta data. In addition to the first example, where we displayed how to fetch a snapshot of an image from PMA.start using Python, we can extend the example slightly so that we can feed the snapshot image to CellProfiler for further analysis.ĬellProfiler can be invoked by the command line and instructed to execute a pipeline against a particular image.
CellProfiler can be retrieved from its own website.


 0 kommentar(er)
0 kommentar(er)
